Iridium »
PDB 1c1k-5de8 »
1c1k »
Iridium in PDB 1c1k: Bacteriophage T4 Gene 59 Helicase Assembly Protein
Protein crystallography data
The structure of Bacteriophage T4 Gene 59 Helicase Assembly Protein, PDB code: 1c1k
was solved by
T.C.Mueser,
C.E.Jones,
N.G.Nossal,
C.C.Hyde,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 5.00 / 1.45 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 31.865, 65.882, 108.166, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 25.3 |
Other elements in 1c1k:
The structure of Bacteriophage T4 Gene 59 Helicase Assembly Protein also contains other interesting chemical elements:
| Chlorine | (Cl) | 12 atoms |
Iridium Binding Sites:
The binding sites of Iridium atom in the Bacteriophage T4 Gene 59 Helicase Assembly Protein
(pdb code 1c1k). This binding sites where shown within
5.0 Angstroms radius around Iridium atom.
In total 2 binding sites of Iridium where determined in the Bacteriophage T4 Gene 59 Helicase Assembly Protein, PDB code: 1c1k:
Jump to Iridium binding site number: 1; 2;
In total 2 binding sites of Iridium where determined in the Bacteriophage T4 Gene 59 Helicase Assembly Protein, PDB code: 1c1k:
Jump to Iridium binding site number: 1; 2;
Iridium binding site 1 out of 2 in 1c1k
Go back to
Iridium binding site 1 out
of 2 in the Bacteriophage T4 Gene 59 Helicase Assembly Protein
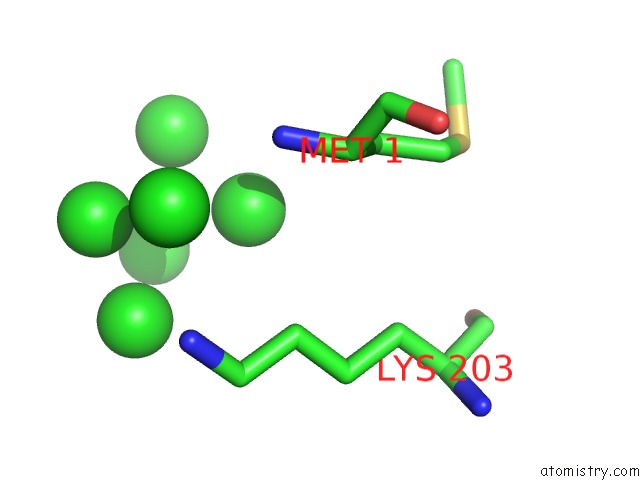
Mono view
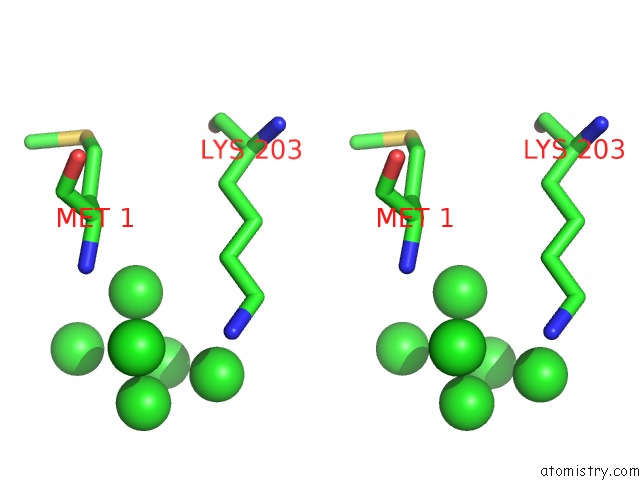
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iridium with other atoms in the Ir binding
site number 1 of Bacteriophage T4 Gene 59 Helicase Assembly Protein within 5.0Å range:
|
Iridium binding site 2 out of 2 in 1c1k
Go back to
Iridium binding site 2 out
of 2 in the Bacteriophage T4 Gene 59 Helicase Assembly Protein
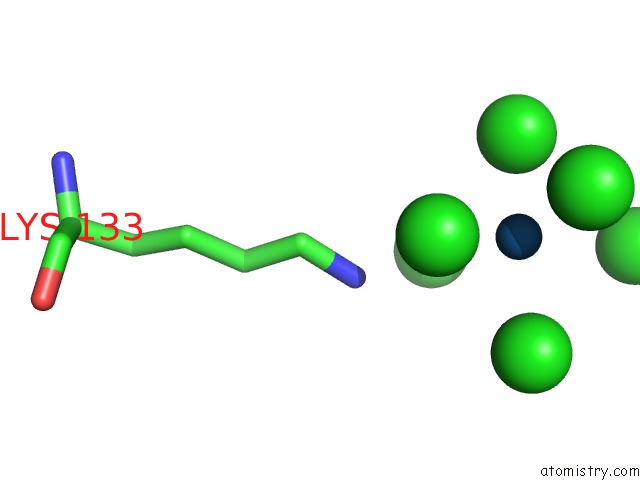
Mono view
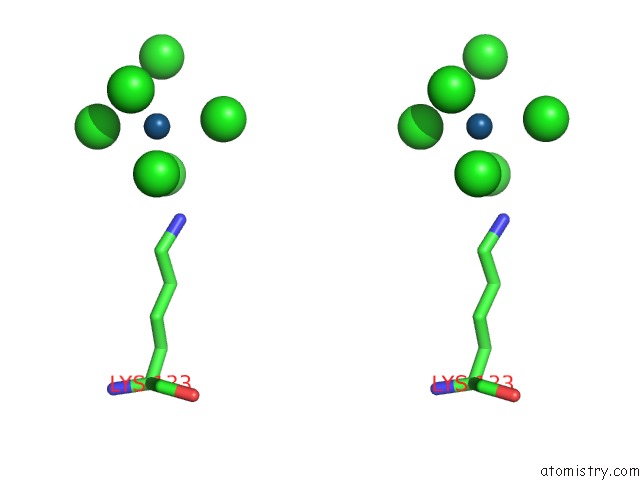
Stereo pair view

Mono view

Stereo pair view
A full contact list of Iridium with other atoms in the Ir binding
site number 2 of Bacteriophage T4 Gene 59 Helicase Assembly Protein within 5.0Å range:
|
Reference:
T.C.Mueser,
C.E.Jones,
N.G.Nossal,
C.C.Hyde.
Bacteriophage T4 Gene 59 Helicase Assembly Protein Binds Replication Fork Dna. the 1.45 A Resolution Crystal Structure Reveals A Novel Alpha-Helical Two-Domain Fold. J.Mol.Biol. V. 296 597 2000.
ISSN: ISSN 0022-2836
PubMed: 10669611
DOI: 10.1006/JMBI.1999.3438
Page generated: Mon Aug 12 03:19:59 2024
ISSN: ISSN 0022-2836
PubMed: 10669611
DOI: 10.1006/JMBI.1999.3438
Last articles
Ca in 2ML2Ca in 2MLF
Ca in 2ML1
Ca in 2MLE
Ca in 2MGU
Ca in 2MES
Ca in 2MKP
Ca in 2MIN
Ca in 2MAS
Ca in 2MBX